New insight into RNA regulation in muscle
Muscle disorders such as muscular dystrophies or heart disease affect thousands of people worldwide. One feature shared by many muscle diseases, as well as neuronal disorders and even cancers, is a change in RNA regulation. Genes, which are encoded in our DNA, have to be copied into an mRNA that can be read by the molecular machines that produce proteins and build our cells. Each type of muscle cell, for example heart or skeletal muscle, produces its own, specific set of mRNAs. In a cell from a patient with a muscle disorder, this set of mRNAs deviates from the one a healthy muscle cell produces, and the wrong protein or the wrong amount of a specific protein is produced. This leads to defects in the construction of the sarcomere mini-motors that power muscle movement, and is thought to lead to symptoms of muscle disease.
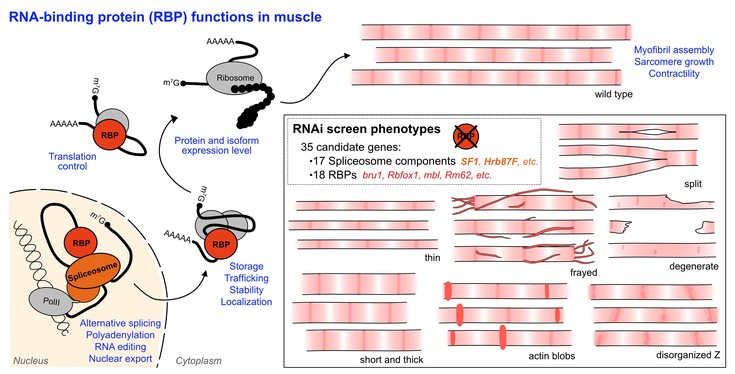
Our cells contain hundreds of regulatory proteins, known as RNA binding proteins, that act like product managers and organize which mRNAs are produced in each cell. Unfortunately, only 10% of these RNA binding proteins have been studied so far in muscle cells, so their role in RNA regulation is not well understood. This makes it challenging to understand how defective RNA regulation leads to disease phenotypes, or in other words which product defects caused by poor management are responsible for disease. A better understanding of RNA regulation and RNA binding proteins will eventually lead to development of new and better treatments for muscle disease.
Dr. Maria Spletter and her team are working to expand our knowledge of RNA binding protein function in muscle. In a new study using the fruit fly Drosophila melanogaster, they show that loss of 35 different RNA binding proteins leads to a wide variety of muscle defects, from diverse flaws in construction of sarcomeres to problems with muscle contraction and complete loss of muscle fibers. These defects depend on how much or how little of an RNA binding protein is present in a muscle cell. Muscles are highly conserved and have a similar structure in fruit flies and humans, so this work provides insight into what these proteins likely do in human muscle.
Dr. Maria Spletter and her team go on to report a novel, muscle-specific role for the protein SF1 in maintaining stability of the sarcomere Z-disc and additionally identify a conserved myogenic role for Hrb87F, an hnRNPA homolog, which has been shown previously to regulate muscle development in mammals. Both SF1 and Hrb87F regulate hundreds of alternative splice events in developing flight muscle. To understand if this splicing program is important to the identity of the muscle, they compared the set of mRNA transcripts found in SF1 and Hrb87F to data on Bruno1. Bruno1 is a fly homolog of the human protein CELF1, a major RNA binding protein misregulated in myotonic dystrophy, which orchestrates a flight-muscle-specific RNA regulatory program. Notably, the targets of Bruno1 are different from the targets regulated by SF1 and Hrb87F, suggesting these two regulatory programs function in parallel. This finding implies that multiple RNA regulatory programs concurrently co-exist in a muscle cell.
The study of Dr. Maria Spletter and team is an important step towards building a more comprehensive understanding of RNA binding protein function in muscle. It also highlights the utility of genetic model organisms such as Drosophila to systematically identify new RNA binding proteins with muscle-specific functions. Dr. Spletter and her team set the stage for further study of the components and complexity of this RNA regulatory network, to understand how muscle-type identity is integrated with more general RNA regulatory pathways in healthy muscle, and how this is disrupted in muscle disease.
Publication: Kao, Nikonova et al: A Candidate RNAi Screen Reveals Diverse RNA-Binding Protein Phenotypes in Drosophila Flight Muscle, Cells 2021