New insights into the dynamic control of DNA damage repair
Our genome contains large amounts of “fragile” regions, where DNA breaks frequently occur. Those fragile sites are often characterized by the presence of repetitive sequences. Repair of these DNA sequences can lead to genetic rearrangements through a repair mechanism, which uses templates from homologous DNA sequences and is therefore known as homologous recombination (HR). Uncontrolled HR is often associated with neurodegenerative diseases and cancer. Therefore, repetitive DNA is often shielded from repair factors by sequestration into specific compartments, such as the nucleolus that contains hundreds of repeats of the ribosomal DNA (rDNA).
But what happens if rDNA repeats are damaged? How can they be repaired? Previous studies have shown that damaged rDNA repeats can be transiently released from the nucleolus to allow access to the DNA repair machinery. However, the molecular mechanism that triggers the release has remained unknown.
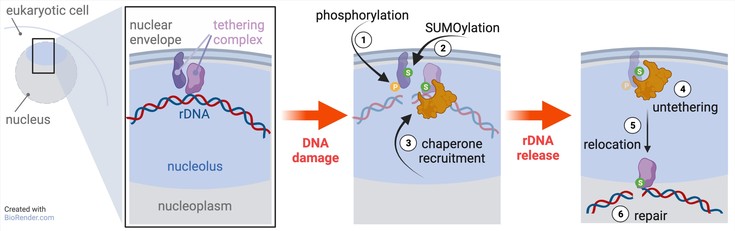
In their latest work, the team of Sigurd Braun unveiled how rDNA repeats are released to the nucleoplasm during DNA damage (Capella et al., 2021) in a collaboration with Andreas Ladurner and Boris Pfander. In the unicellular model organism budding yeast (Saccharomyces cerevisiae), the rDNA repeats are tethered to the inner nuclear membrane and sequestered inside the nucleolus through the interaction of two protein complexes, which together form the rDNA tethering complex. Using a combination of biochemistry, microscopy and elegant yeast genetics, Capella et al. showed that multiple steps trigger the rDNA release from the nuclear envelope and into the nucleoplasm. First, DNA damage results in the phosphorylation of the nuclear membrane complex. This modification promotes the addition of another molecular ‘tag’ known as Small Ubiquitin-like Modifier, or SUMO, on both protein complexes involved in tethering. Proteins marked by SUMO are recognized by distinct molecular machineries, for instance the ATP-dependent chaperone Cdc48 (p97 in mammals). Through specific recognition motifs, Cdc48 binds to the SUMOylated tethering complex and breaks up the interaction between its partners in an ATP-dependent manner, resulting in rDNA release from the nucleolus and DNA repair by HR machinery. Importantly, Capella et al. found that nucleolar rDNA release in mammalian cells also requires the activity of Cdc48/p97, implying that this repair mechanism is conserved in higher eukaryotes.
Together, the findings by Capella and co-workers provide crucial insights into the dynamic regulation of DNA damage repair. Noteworthy, while yeast cells deficient in rDNA tethering show hyper-recombination and loss of individual rDNA repeats reminiscent of cancer, the authors found that cells that cannot untether rDNA from the nuclear envelope are not viable. This implicates that rDNA untethering happens frequently in eukaryotic cells, illustrating the importance of this regulatory mechanism.
Publication: Capella et al.: Nucleolar release of rDNA repeats for repair involves SUMO-mediated untethering by the Cdc48/p97 segregase, Nature communications 2021

